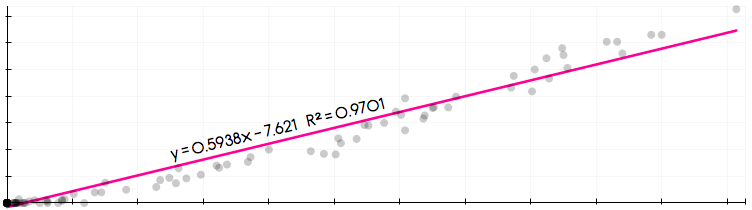
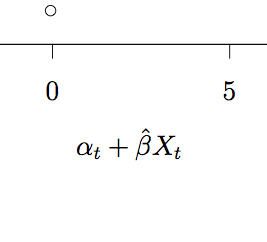The following code will attempt to replicate the results of the numpy.linalg.lstsq() function in Numpy. For this exercise, we will be using a cross sectional data set provided by me in .csv format called “cdd.ny.csv”, that has monthly cooling degree data for New York state. The data is available here (File –> Download).
The OLS regression equation:
where a white noise error term. For this example
the population-weighted Cooling Degree Days (CDD) (CDD.pop.weighted), and
CDD measured at La Guardia airport (CDD.LGA). Note: this is a meaningless regression used solely for illustrative purposes.
Recall that the following matrix equation is used to calculate the vector of estimated coefficients of an OLS regression:
where the matrix of regressor data (the first column is all 1’s for the intercept), and
the vector of the dependent variable data.
Matrix operators in Numpy
matrix()coerces an object into the matrix class..Ttransposes a matrix.*ordot(X,Y)is the operator for matrix multiplication (when matrices are 2-dimensional; see here)..Itakes the inverse of a matrix. Note: the matrix must be invertible.